Parallel tempered Markov Chain Monte Carlo sampler.
More...
#include "sampling/sampler_differential_evolution_tempered_MCMC.h"
|
|
| sampler_differential_evolution_tempered_MCMC (int seed) |
| | Class constructor, accepts the integer seed for a random number generator as an argument.
|
| |
| void | run_sampler (likelihood _L, int length, int temp_stride, int chi2_stride, std::string chain_file, std::string lklhd_file, std::string chi2_file, std::vector< double > means, std::vector< double > ranges, std::vector< std::string > var_names, bool continue_flag, int output_precision=6, int verbosity=0, bool adaptive_temperature=true, std::vector< double > temperatures=std::vector< double >(0)) |
| | Function to run the sampler, takes a likelihood object, name of output files, and tuning parameters for the sampler and returns the MCMC chain, likelihoods and chi squared values. More...
|
| |
|
double | RndGaussian (double, double, bool) |
| | Function to generate random numbers with a gaussian distribution.
|
| |
|
double | RndUni (double, double) |
| | Function to generate real valued random numbers in an interval.
|
| |
|
int | RndUnint (int, int) |
| | Function to generate integer valued random numbers in an interval.
|
| |
| void | set_cpu_distribution (int num_temperatures, int num_walkers, int num_likelihood) |
| | Function to set the distribution of processors in different layers of parallelization. More...
|
| |
|
void | set_tempering_schedule (double t0, double nu=1.0, double T_ladder_factor=5.0) |
| | Function to set the parameters of the tempering schedule. t0 is the halving-time, nu is the origional tempering value, which should be bounded by unity.
|
| |
| void | set_checkpoint (int ckpt_stride, std::string ckpt_file) |
| | Function to set the checkpoint/restart functionality. More...
|
| |
| void | estimate_bayesian_evidence (std::vector< std::string > file_names, std::vector< double > temperatures, int burn_in) |
| | Function to estimate the bayesian evidence using the Thermodynamic Integration method. More...
|
| |
|
std::vector< double > | find_best_fit (std::string chain_file, std::string lklhd_file) |
| | Finds the best fit within provided chain file and returns it.
|
| |
Runs parallel tempered differential evolution ensemble sampling Markov Chain Monte Carlo chains to sample the likelihood surface. This routine uses parallel tempering on top of the differential evolution method of Cajo J.F Ter Braak (2006). This implementation closely follows that of B. Nelson et. al (2013) used for analyzing radial velocity observations (RUN DMC code). Parallel tempering is optimized by dynamically adjusting the temperature ladder as described in W. D. Vousden et. al (2016). Given an object of type likelihood (which encompases the likelihood, priors and chi squared) the sampler explores and samples the likelihood surface over its dependent parameters. It will provide a sampling of the posterior probability distribution. The likelihood and the chi squared evaluated at the sampled points are also provided by the sampler.
| void estimate_bayesian_evidence |
( |
std::vector< std::string > |
file_names, |
|
|
std::vector< double > |
temperatures, |
|
|
int |
burn_in |
|
) |
| |
- Parameters
-
| file_names | Vector of strings holding the names of the likelihood files corresponding to each tempered level. |
| temperatures | Vector containing the temperature values for the corresponding log-likelihood sample files. The order of temperatures and likelihood file names in their vectors must be the same. |
| burn_in | Number of burn-in samples to exclude from the analysis. |
| void run_sampler |
( |
likelihood |
_L, |
|
|
int |
length, |
|
|
int |
temp_stride, |
|
|
int |
chi2_stride, |
|
|
std::string |
chain_file, |
|
|
std::string |
lklhd_file, |
|
|
std::string |
chi2_file, |
|
|
std::vector< double > |
means, |
|
|
std::vector< double > |
ranges, |
|
|
std::vector< std::string > |
var_names, |
|
|
bool |
continue_flag, |
|
|
int |
output_precision = 6, |
|
|
int |
verbosity = 0, |
|
|
bool |
adaptive_temperature = true, |
|
|
std::vector< double > |
temperatures = std::vector<double>(0) |
|
) |
| |
- Parameters
-
| _L | An object of class likelihood. |
| length | Number of steps (stretch moves) taken by the ensemble sampler. |
| temp_stride | Number of steps between subsequent communication among chains of different temperatures. |
| chi2_stride | Number of steps between outputing Chi squared values. |
| chain_file | String variable holding the name of the output MCMC chain file. |
| lklhd_file | String variable holding the name of the output likelihood file, contains log-likelihood values for each MCMC step. |
| chi2_file | String variable holding the name of the output chi squared file, contains chi squared values for the MCMC chain. |
| means | Vector holding the mean values of parameters used for initializing the MCMC walkers. |
| ranges | Vector holding the standard deviation of parameters used for initializing the MCMC walkers. |
| var_names | A vector of strings. It holds the names for each sampled variable. The names are compiled as a header in the "chain_file". If the vector doesn't contain any names the header will not be generated. If the header is present The Themis analysis tools can use it to correctly label the generated diagnostics plots. |
| continue_flag | Boolean variable. If set to "True" the sampler would use a checkpoint file to resume it's state and continue the run. If the output files exist the new data is appended to the same files. If set to "false" it will start a new chain using the provided "means" and "ranges" variables to initialize the chain. Note in the latter case existing output files will be overwitten by the new ones. |
| output_precision | Sets the output precision, the number of significant digits used to represent a number in the sampler output files. The defaul precision is 6. |
| verbosity | If set to one chain files will be produced for all tempering levels, otherwise only the lowest temperature will produce a chain file which is the deisred posterior probability distribution |
| adaptive_temperature | If set to "true" the code will iteratively adapt the temperature ladder to get optimize the parallel tempering. If set to "false" the temperatures would remain constant. The latter case can be useful if one needs to find the bayesian evidence from the output postriors/likihoods at fixed temperatures. The default setting is "true" which is the best choice for most cases. |
| temperatures | Optional vector to set the temperatures used for parallel tempering. |
| void set_checkpoint |
( |
int |
ckpt_stride, |
|
|
std::string |
ckpt_file |
|
) |
| |
- Parameters
-
| ckpt_stride | Number of steps between writing a new checkpoint. |
| ckpt_file | String variable holding the name of the output checkpoint file. |
| void set_cpu_distribution |
( |
int |
num_temperatures, |
|
|
int |
num_walkers, |
|
|
int |
num_likelihood |
|
) |
| |
- Parameters
-
| num_temperatures | Integer value ( \( \geq 1 \)). Number of temperatures used by the parallel tempering algorithm. If set to one, the sampler will run without tempering. |
| num_walkers | Number of walkers used by ensemble sampler. This should be at least a few times the dimension of the parameter space. |
| num_likelihood | number of threads allocated for each likelihood calculation. |
The following plot shows how the sampler scales with different number of walkers per MPI process. The green line shows the ideal case of linear scaling where the run time is inversely proportional to the number of MPI processes used. The purple line shows how the sampler scales with the number of MPI processes. As can be seen in the figure the scaling closely follows linear scaling and always remains within \(\%20\) of the ideal linear scaling.
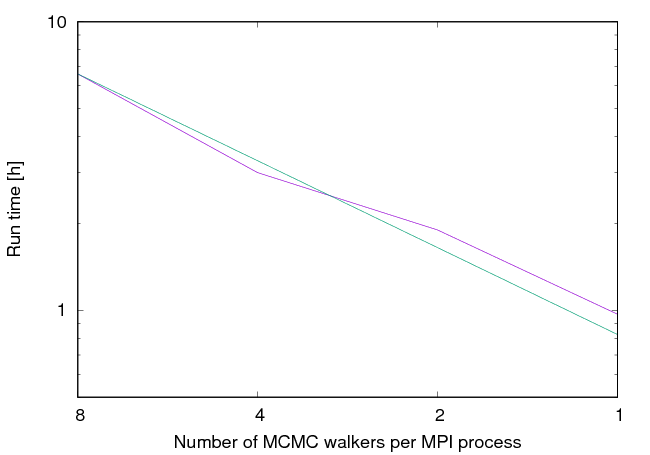
Sampler scaling plot. The green line shows the linear scaling.
The documentation for this class was generated from the following files: