JupyterHub on Symmetry
To log in, visit https://symmetry.pi.local. As you might guess from the name, this is currently only available within the Perimeter network. (If you want to access it from outside, please use the PI VPN.)
Currently, you will get a warning that Your connection is not secure, because we don’t have a proper security certificate yet. Please tell your web browser that this is okay (in Firefox: Advanced → Add Exception → Confirm Security Exception).
Log in using your normal PI username and password, without the
@perimeterinstitute.ca.
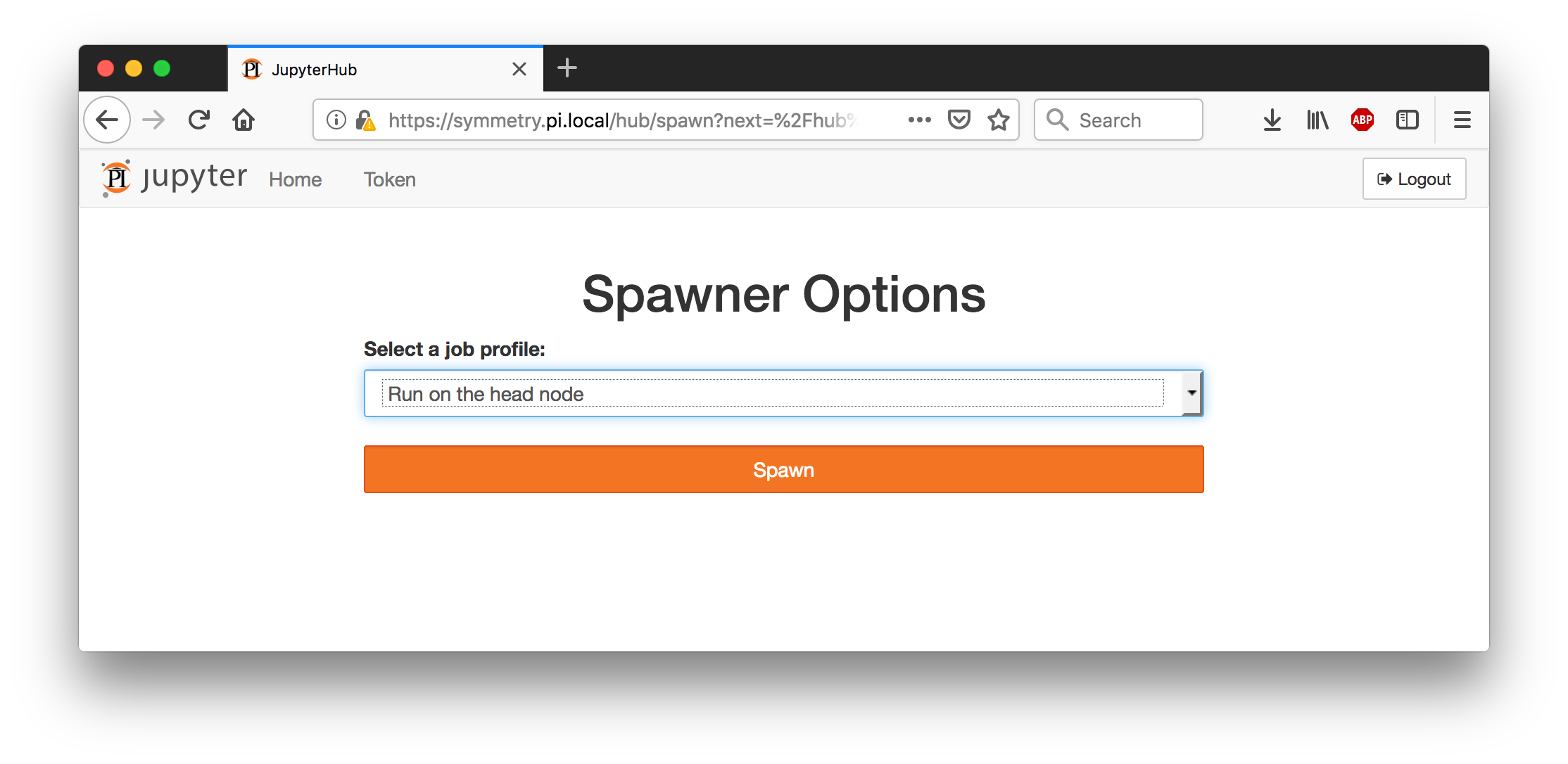
You will arrive at a Spawner Options page with a drop-down menu,
asking you where you would like to run your Jupyter notebook. You can
run on the head node—a computer shared with many other users—or on
one of the compute nodes. The head node will be available
immediately, while the compute nodes may take a while to become
available, because we have to wait for a batch job to be scheduled.
The compute nodes are more powerful and are more suitable for
expensive computations or memory-hungry applications.
Once you have selected where to run your notebook, you will arrive at the main Jupyter notebook page, which by default will show you your home directory. From here, you can Upload files (using the Upload button at the top-right), and download files (by clicking on them). At the top-right there is a New drop-down menu to start a new Jupyter kernel, an interactive programming environment.
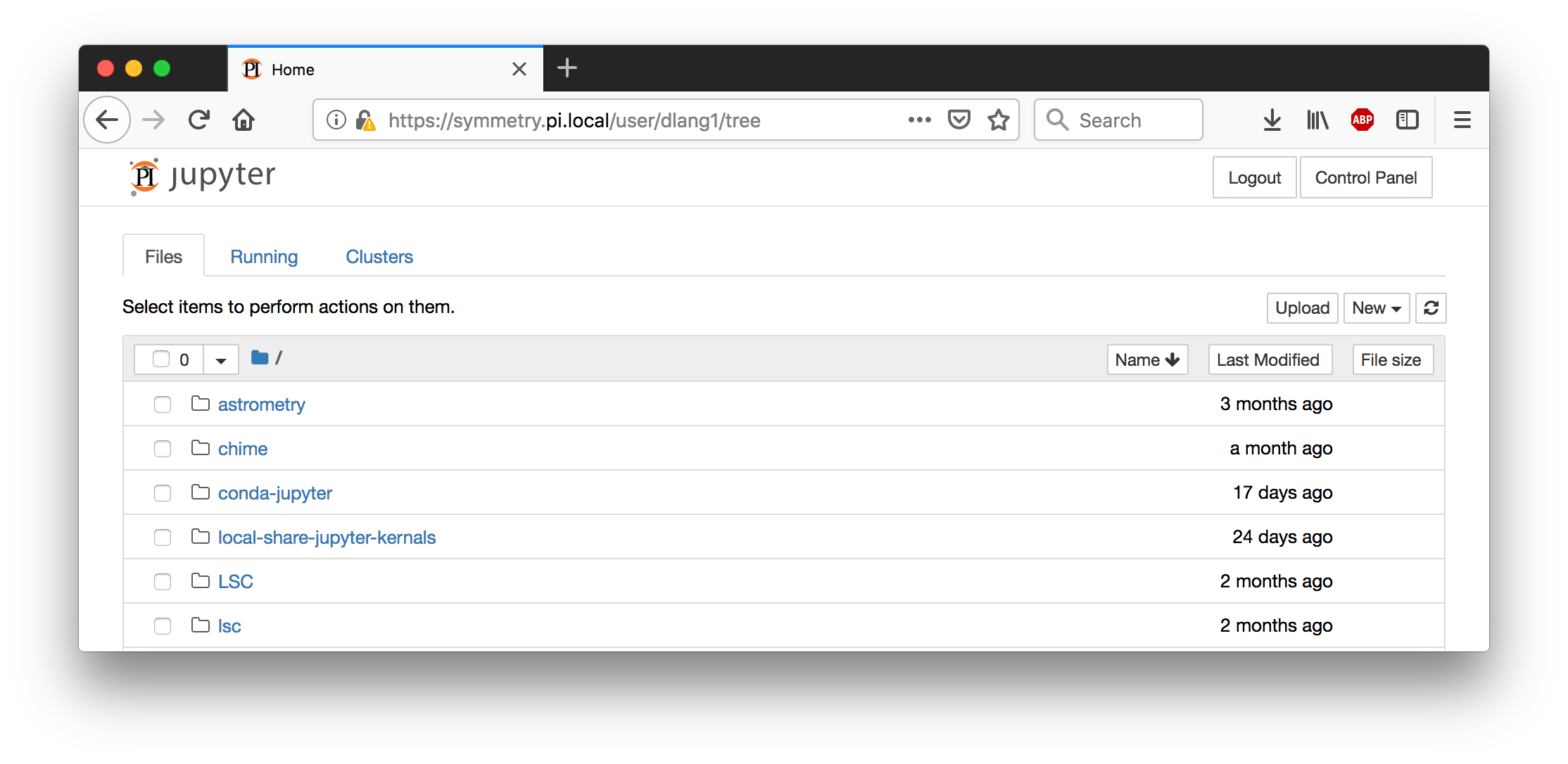
If you started running on the head node but would now like to run on a compute node, use the Control Panel button at the very top-right, then hit the Stop My Server button, then the Start My Server button will take you back to the Spawner Options page.
Currently, we have several Python versions, a Bash (shell) environment, and a recent Julia version. If you want to use a different programming language, please write a note to help@perimeterinstitute.ca with the details!
Julia
Before using a Julia kernel through Jupyterhub, unfortunately, you must
install the IJulia package using the Julia command line:
$ module load julia
$ julia
_
_ _ _(_)_ | Documentation: https://docs.julialang.org
(_) | (_) (_) |
_ _ _| |_ __ _ | Type "?" for help, "]?" for Pkg help.
| | | | | | |/ _` | |
| | |_| | | | (_| | | Version 1.1.0 (2019-01-21)
_/ |\__'_|_|_|\__'_| |
|__/ |
julia> using Pkg
julia> Pkg.add("IJulia")
Cloning default registries into `~/.julia`
Cloning registry from "https://github.com/JuliaRegistries/General.git"
...
We have not figured out a good way to provide the IJulia package
(which connects Julia to Jupyterhub) on a system-wide basis, so this
installs the IJulia package in your home directory.
Haskell
It is possible (but not super easy) to set up a Haskell kernel. If you are
interested in doing this, please email help@perimeterinstitute.ca.
Custom Kernels
It is possible to add your own kernels to the list of kernels. When it starts, Jupyterhub will search for kernel files in these directories:
/cm/shared/apps/conda-environments/jupyterhub/share/jupyter/kernels/
$HOME/.local/share/jupyter/kernels
To create a new kernel for yourself (say, using a special Conda environment you have created), you can copy one of the existing kernel files and modify it for your needs. For example:
mkdir -p ~/.local/share/jupyter/kernels
cp -r /cm/shared/apps/jupyter-kernels/python37-conda ~/.local/share/jupyter/kernels/mykernel
# Edit ~/.local/share/jupyter/kernels/mykernel/kernel.json
# to point to the "python" executable inside your environment.